Proteomics
The TSL proteomics team map post-translational modifications and uncover protein–protein interactions underlying host–microbe interactions.

Our Research
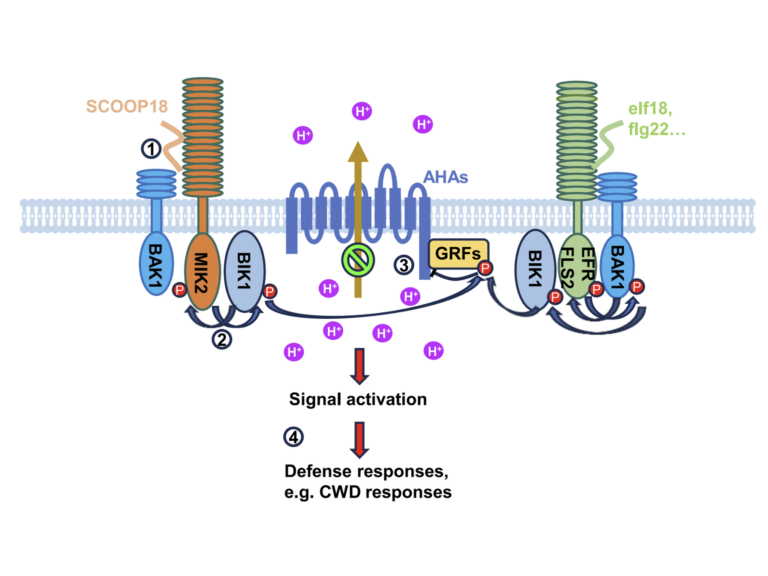
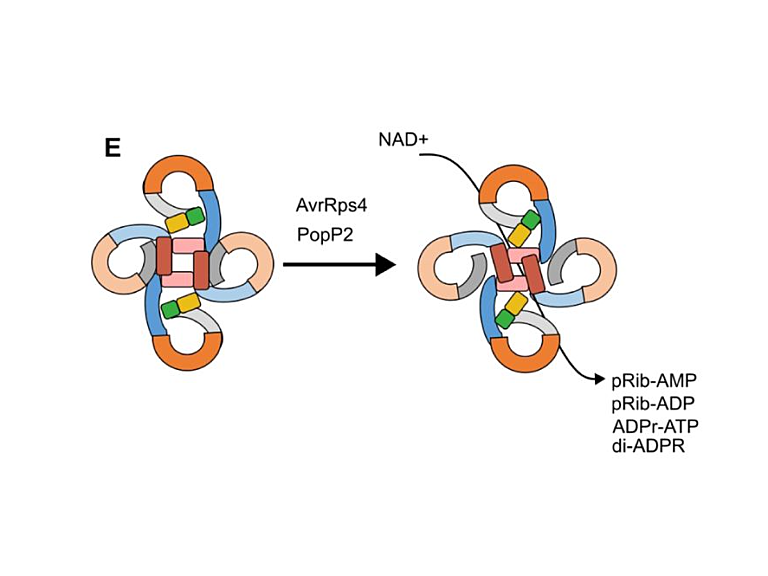
Mapping post-translational modifications on the proteome and identifying protein-protein interactions in the context of host microbe interactions remains a formidable challenge, despite the resolving power of the latest mass spectrometers. The proteomics team at TSL addresses this challenge through collaborative projects using our discovery proteomics platform.