Publications

TSL scientists publish their work in a wide range of academic journals.
We are strong supporters of open access science and open access publishing. TSL is a signatory of the San Francisco Declaration on Research Assessment (DORA; https://sfdora.org/) and actively encourages researchers to use prepublication peer review (preprint servers) and publish in Open Access journals. We publish preprints regularly on bioRxiv and other preprint servers.

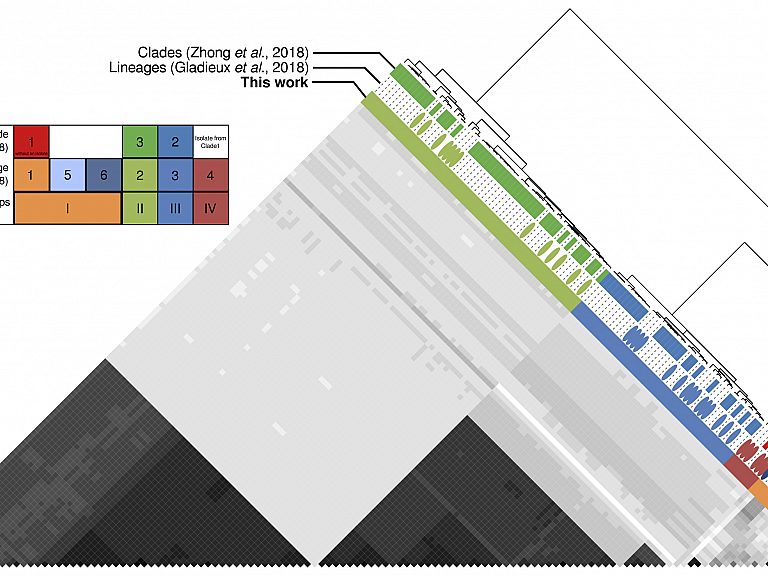
Differential loss of effector genes in three recently expanded pandemic clonal lineages of the rice blast fungus.
Read more
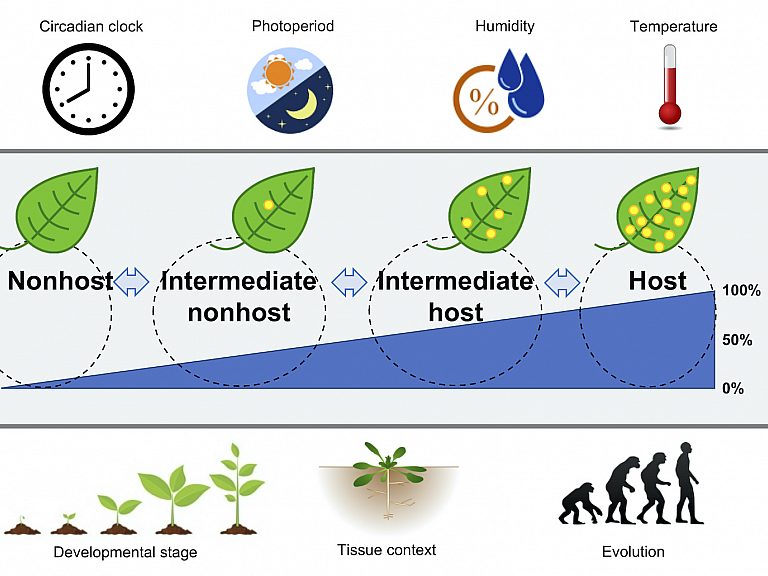
What is the molecular basis of nonhost resistance?
Read more
Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants
Read more
Traffic of a viral movement protein complex to the highly curved tubules of the cortical endoplasmic reticulum
Read more
Functional characterization of a gene family encoding polygalacturonases in Phytophthora parasitica
Read more